Home
Home
- AWG Runs
- Contact Us
- CPTAC Meeting Agendas & Notes
- Dashboard-Analyses
- Dashboard-Stddata
- Data Usage Policy
- Documentation
- Download
- FAQ
- fbget
- FireBrowse Release Notes
- FireCloud Feature Requests
- GDAN_GDC_Notes
- Legacy
- MAF Dashboard
- Mission Statement
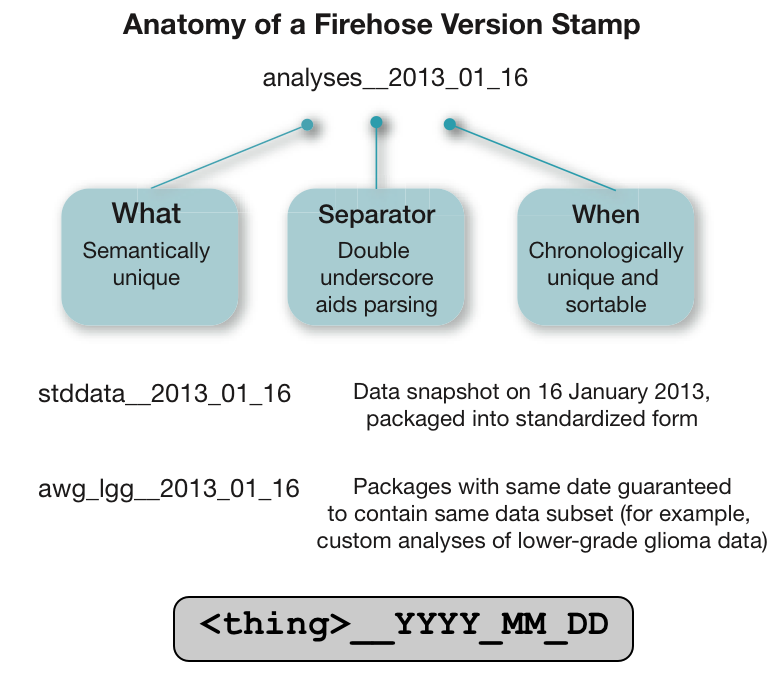
- Nomenclature
- Nozzle
- Presentations
- ProcessFlow
- QualityControl
- Rationale
- VizDev Team Meeting Notes
As of December 2013 this is no longer the public-facing home page for the Broad GDAC Firehose pipeline. Please visit gdac.broadinstitue.org for the Broad GDAC Firehose home page.
Born of the desire to systematize analyses from The Cancer Genome Atlas pilot and scale their execution to the dozens of remaining diseases to be studied, Firehose now sits atop ~40 terabytes of TCGA data and reliably executes more than 6000 pipelines per month.
The Broad Institute TCGA GDAC Firehose Provides
Version-stamped, standardized datasets
Precursor to automated analyses: aggregates all available sample batches into a single, uniformly-formatted bolus (one per disease X datatype), which can be immediately fed to algorithmic codes without further data preparationVersion-stamped packages of standard scientific analysis results
Automatically generated for dozens of algorithms: GISTIC, MutSig, Clustering, Correlation, ...Version-stamped, biologist-friendly reports
Encapsulating analysis results in a form accessible to a wide audience, online for public browsing, and citable in the literature through DOIsVersion-stamped custom runs for TCGA analysis working groups
Performed by request in support of TCGA marker paper analysis, on a much shorter timescale than the monthly data runs and quarterly analysis runs.
These can be explored & retrieved interactively through our data dashboard and analysis dashboard, or downloaded en masse with firehose_get.Towards the aim of reproducibility, our online suite of reports provides thousands of pages of documentation for the analyses performed; in addition, extensive release notes are available for each versioned dataset and analysis package release. Finally, more information is available in many of the talks and posters we've presented and our online FAQ.
For a discussion of Firehose in the broader context of Big Cancer Data, see Nature Methods 10, 293–297 (2013) doi:10.1038/nmeth.2410