As of 2017, our archives follow the new nomenclature given below:
<DiseaseCohortName>.<TaskName>.<RunCode>.<Revision>
Element | Description of Permissible Values | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
DiseaseCohortName | A string of the form <project_name>-<disease_specification>[-<sample_type>] for example: TCGA-ACC-TP. The <disease_specification> most often refers to a single disease study given by its disease abbreviation , such as GBM for Glioblastoma Multiforme; but may also refer to an aggregate of multiple diseases, such as PANCAN12 (which refers to a cohort of 12 diseases created to study pan-cancer trends) or COADREAD (which combines the single diseases COAD and READ into one cohort). The optional <sample_type> suffix consists of a literal dash followed by a sample type code designating the tissue sample type; for example, the suffix "-TP" indicates that the given archive contains results based upon primary tumor data. As a final example, here's how sample type codes would most commonly map to sample sets in Firehose, for a single disease study:
| ||||||||||||||
TaskName | Tasks should be named as <Datatype>_<AlgorithmName> For example: CopyNumber_Gistic2. The datatypes correspond to columns 2-12 in any of our sample data tables with several types spelled out in longer form for clarity as follows:
| ||||||||||||||
RunCode | Eight numeric characters representing the date. For example, 20170807 indicates August 7, 2017. | ||||||||||||||
Revision | A small integer (usually single digit) indicating how many times the given <TaskName> was successfully run in the given pass. |
Legacy TCGA (prior to 2017)
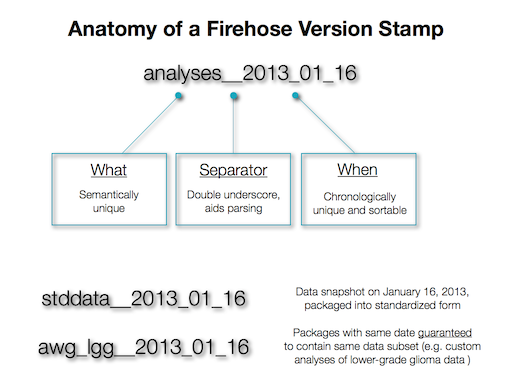
Each pipeline executed by the BROAD TCGA GDAC Firehose pipeline results in a set of 6 files being submitted to the DCC: primary results in the Level_* archive; auxiliary data (e.g. debugging information) in the aux archive, tracking information in the mage-tab archive; and an MD5 checksum file for each. In most cases you will only need the primary results in the Level_* archives. Microsoft Windows-based users can use the WinRAR utility to unpack the archive files, while Unix and Apple Mac OS/X users can use the gzip and/or tar utilities. As of January 2013 our archives follow the nomenclature given below. Look here for older version.
<Domain>_<DiseaseCohortName>.<TaskName>.<DataLevel>.<Runcode>.<Revision>.0
Element | Description of Permissible Values | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Domain | the literal string gdac.broadinstitute.org | ||||||||||||||
DiseaseCohortName | a string of the form < The disease specification most often refers to a single disease study given by its TCGA disease abbreviation, such as GBM for Glioblastoma Multiforme; but may also refer to an aggregate of multiple diseases, such as PANCAN12 (which refers to a cohort of 12 diseases created to study pan-cancer trends) or COADREAD (which combines the single diseases COAD and READ into one cohort). The optional <sample_type> suffix consists of a literal dash followed by a TCGA short letter code designating the tissue sample type; for example, the suffix "-TP" indicates that the given archive contains results based upon primary tumor data. As a final example, here's how TCGA short letter codes would most commonly map to sample sets in Firehose, for a single disease study:
| ||||||||||||||
TaskName | Tasks should be named as <Datatype>_<AlgorithmName> For example: CopyNumber_Gistic2. The datatypes correspond to columns 2-12 in any of our sample data tables with several types spelled out in longer form for clarity as follows:
| ||||||||||||||
DataLevel | the literal strings Level_2 or Level_3 for stddata tasks, or Level_4 for analyses tasks | ||||||||||||||
Runcode | 10 alphanumeric characters representing the date and a unique "pass" identifier, such as 2011072800 to indicate "pass 0" over the July 28,2011 data snapshot; or 2011072801 to indicates "pass 1" over same dated snapshot | ||||||||||||||
Revision | a small integer (usually single digit) indicating how many times the given TaskName was successfully run in the given pass |