CPTAC Meeting Agendas & Notes
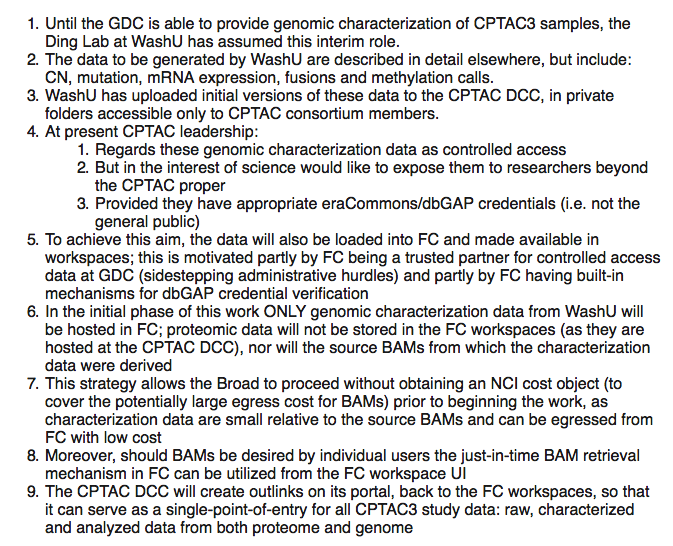
Oct 29, 2018:
Just a reminder that PDC has issued an alpha of it's MVP release. Would be nice to have more Broadies take a look. Presently supports only CPTAC2 data.
- iCoMut:
- undergoing update for UCEC AWG
- FYI: HG38 mutation calls underway by WashU (slated for 11/8 V2 freeze)
- waiting for Broad example data to add new proteomics tracks
- undergoing update for UCEC AWG
- HG38: progress, GDCtools config file needed
- CN data:
/xchip/gdac_data/cptac3/genomic_data_mirror/CPTAC3_WashU_hg38/UCEC/WXS_CNV_hg38_v1.0_20180809
But what is more biologically important: WGS for CN or HG38?
Because at present only VCFs available for WGS CN data
Feedback: Mani says WES is fine for Broad PG pipeline
Karl needs WGS for some of his investigatory work (non-canonical ORFs, from HLA), but that's not ready for prime time sharing with other PGDACs yet, so it doesn't impact our G-CDAP
- CN data:
- LUAD: new proteomic data "in progress," but in "new location" @ DCC:
- per this email exchange
Changed from/to:
8_CPTAC3_Lung_Adeno_Carcinoma/LUAD_mzML/
8_CPTAC3_Lung_Adeno_Carcinoma/A_CPTAC3_hg38_CDAP/LUAD_mzML/
Per Nathan: "The files under A_CPTAC3_hg38_CDAP represent the re-run of the CDAP
analysis for files analyzed before the release of Broad's new
RefSeq/hg38 protein sequence database and any new ones going forward."
Sept 17, 2018:
Upcoming milestones:
- TCGA Legacy symposium (9/26-9/29, posters starting this week)
- CPTAC F2F (Oct 15-18):
- for F2F our only licensing concern is access to the CPTAC communit
- so each user who wants access sends email to us, and is granted, b/c they are members of CPTAC consortiumn
- Paper on proteomics pipeline (Dec 2018)
These all imply that pipelines should be made public-ready in near future
- Definition: releasing pipeline "in FC" is treated effectively as releasing source code
- In part because docs usually live in repo wiki, and so it must be opened to public for (unless docs are duplicated to elsewhere)
- Do we need to have an "I Agree" dialogue box for codes which require licenses? For example, on codes which come from other labs.
- At the very least, for EXTERNAL codes used in our pipeline OUR codes which need commercial tools (like MatLab REs)
- For proteomic pipeline, Mani says it's ALL OK, everything can be run, for whatever reason, by anyone
- But we need to go over EVERY task in GDAC genomic pipeline to ensure all components (especially external codes) are releasable
- Including APOBEC, GSEA (and also tools like Philosopher from proteomic pipeline)
- We may want to release "genomic lite" pipeline, with certain tools omitted
- Possible interim solution: until "I Agree" is possible at runtime in FC, to prevent commercial entities from profiting:
- Karl made strong point about re-distribution being what most method developers want to control/prevent, but how?
- Mike pointed out that while preventing re-distrib is important, mere "runnability" also needs to be monitored/protected:
- otherwise commercial entities can take our labor/tools (or even re-brand as their own) & then profit by analyzing/etc their data
- Chet pointed out that GenePattern team wrestled with this years ago, eventually taking the approach: "We can't enforce good behavior," but we can record whether LICENSE terms have been acknowledged & record in a database, as well as record of each instance when a any user ran any tool. Then let the lawyers fight it out if need be.
- Mike suggested that enforcing acknowledgement of LICENSE terms can be done with a boolean attribute or config parameter, which defaults to False, but user sets to True to signify that LICENSE terms have been read and/or accepted. This facilitates "I Agree" dialog box suggested above for FireCloud UI, but also covers the programmatic use case.
Is it time for us to Hydrant-ize our GDAN configs, which would benefit both genomic & proteomic pipelines (GDAN & CPTAC)
Mani & Karsten considered this for proteo-genomic portion of pipelines, and decided NO, not until after Oct 2018 F2F (and possibly Dec paper)
CGA then adopted similar position.
- Prioritization:
- This is substantial effort: really need extra SWE effort
- Also: we do not have staffing to respond to wide-ranging GISTIC, MutSig etc user questions
- Lastly: policy on sharing methods/workspaces with collaborators, e.g. Kwiatkowski lab
- if the pipeline is instantiated in our GDAN (or CPTAC workflow)
- and we provide access only through FireCloud
- then the method (and/or method configuration) is essentially for public consumption
- and may be shared through FC with essentially anyone?
- but, in particular, Broadie collaborators (as a first step)
August 6, 2018
Budget alert: 50% gone?
- Methylation data progress
- SOP slides
- Anybody here of "persistently slow Broad institute portal access" from abroad, e.g. China?
- Action: reconnect with PDC, to see
- current progress towards v1 alpha release (Fall 2018)
- chat about when will ingest occur for CPTAC3 data?
June 11, 2018
Latest data?
Progress report: content due soon
- Upcoming site visit: July 17, can mostly come from expanded length/depth of F2F presentation (which was abbreviated to only 10mins)
May 7, 2018
Recap of last week's F2F:
Good FireCloud workshop attendance & feedback
Proteomic:
Genomic:
GDC has made progress automating their pipelines
CPTAC genomic data will be HG38 going forward;
Phased delivery CCRC & UCEC first, to allow publications to be in pipeline by next funding cycle (Gantt chart)
- Two special journals in Fall 2018:
- Special issue Mol. Cell Proteomics:tools, algorithms & computational methods
- Submission deadline will be in November, 2018
- Submission deadline will be in November, 2018
- Journal of Proteome Research: Software Tools and Resources
- Submission deadline September 14, 2018
- Special issue Mol. Cell Proteomics:tools, algorithms & computational methods
- FOA: Sustained Support for Informatics Resources for Cancer Research and Management (U24)
- Due June 14
- Submit LOI by May 14
- Letters of support: gathering now
- Time Permitting: genomic pathway analyses
- GDAN Lung pathifier analyses
- iCluster: Hailei
April 2, 2018
- Access to the GDAC bucket for reference files
- egress pay: need to turn on requestor_pays bit
- authorization domain
- proxy groups and how to keep track of them in a bucket: currently have 2 proxy groups
- CPTAC3 data in FC may help entice new CPTAC users, but it is also akin to replicating DCC
- so, let's wait until potential CPTAC users make explicit requests
- TODO:
- Mani will explore using auth domain for new CPTAC FC users
- Mike will ping NCI about FireCloud SW session in May F2F
- Mike/Sam will price physical hardware & compare to Google VMs, as potential spend for $25K FC disbursement
March 19, 2018
- Review draft agenda for May F2F
- Decide additional attendees
- (Fire)Cloud costs for CPTAC-wide usage: $25K seems to have effectively been reduced
Batches 1 and 2 of genomic data are located at /xchip/gdac_data/cptac3/genomic_data_mirror
WashU has apparently re-submitted batch1 RNASeq data, using MapSlice to map transcripts to gene level
So we should be able, in principle, to proceed with our mRNA pipelines
- Mike has a short, unifying wrapper to all 3 of the DCC upload/download utilities, and can install to Unix server upon request
- Integrative Analyses:
- Karsten: proteomic pathways ...
- Mani: map genomic CN data to LINCs, correlate w/ RNAseq signatures?
- Possibly: multi-omics clustering
- TODO:
- Firecloud workshop scheduling
- Mike push cptac wrapper script to Unix servers
March 5, 2018
Summation of HG19 WashU/GDC workaround & potential recommendations to CPTAC leadership & collaborators.
- Consider: combing the GDC website to see if Dockers are available for their pipelines, and whether these could be instantiated in FC
- Consider: running local MOAP-style pipeline on WXS data, to generate CN, mutation, RNASeq
- Decision: wait for now, it's not fully baked yet
- TODO:
- open edit permissions (on this page) to all viewers
- Identify 2-3 integrative proteo-genomic analyses: but must be on CPTAC3 data
- Combined into iCoMut output
- Planning F2F in May 1,2,3:
- Quilts in FC?
- Although CustomProDB (from Baylor/Bing Zhang group) does similar as Quilts and is already in FC (from Karsten)
- FireCloud workshop
- Karsten & Mani: current instantiation of proteomic pipeline
- On prospective BRCA data
Update to Jan 22, 2018 entry:
Genomic data for CPTAC3 downloaded to: /xchip/gdac_data/cptac3/2018_02_02_genomic_data
- Only 2 cohorts (CRCC and UCEC, i.e. kidney and endometrial) have genomic data available so far
- The 3rd cohort (LUAD, lung) proteomic data not submitted by Broad yet, so WashU has apparently not processed the genomic either
Jan 22, 2018
- Brief review of items missed from last meeting
- Proteogenomic Data Commons Steering Committee:
- Held 2nd advisory meeting call last Wed
WashU/GDC workaround: summary of discussion & decisions from 1/19 call
- New science: degradome?
Jan 8, 2018
- Welcome Yifat Geffen, newest member of CGA
- Brief review of latest suite of genomic run reports (total of 830)
- Whither pathology image browser in CPTAC? The GTEX pathology browser was authored here (and we have strong knowledge of cancer path viewer), so we have a good deal of expertise & code that could in principle be leveraged. I've drafted a suggestion for PAAD dwg here.
- NMF clustering module question (auto-selection of K) from Mani?
- FireCloud hosting of CPTAC data (as partial workaround to lack of CPTAC genomic data at GDC)
- Medblast paper
Dec 11, 2017
- Items from 11/27 meeting that was cancelled
- GDC and CPTAC: summary notes from week of 2017_12_06
- Original plan (and data products) given here
- Impact to CGDAC (the CGA part of proteomics GDAC) sketched below
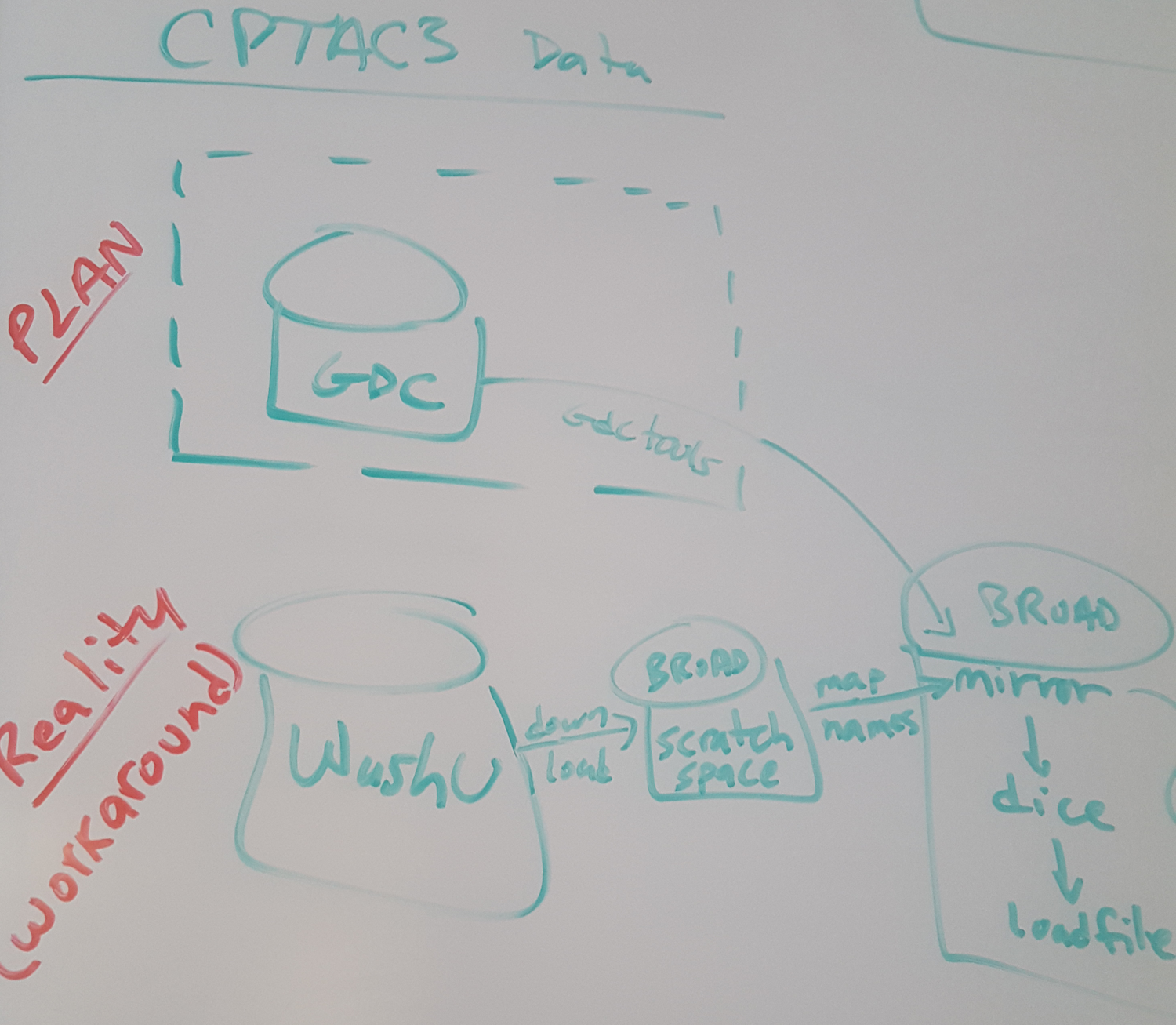
Initial data generation will be shifting from GDC to WashU
Mutation calls (both WES and WGS)
CN generation
RNAseq calls
WashU products deposited to Georgetown DCC
Broad download & remap names as needed/appropriateWhat's next?
FireCloud (as a trusted partner) now being considered as a distribution point
Per Chris Kinsinger feeler conversation on 2017_12_01So, because these data will be HG19 ... our CGA/GDAC in CPTAC may be better utilized by shifting gears, from running existing FireCloud HG38 genomic pipelines on HG19 data (which lead to broken results) ... to loading these HG19 data products from WashU into FireCloud so that it can serve as a distribution point
Side Q: why Georgetown DCC not considered for this? Scale? Absence of trusted partner status?
- Status on $25K to fund use of FireCloud across entire CPTAC?
- any progress: NO, there was an attempt to issue as AWS credits ... currently stuck w/r/t GoogleCredits ... stay tuned
- billing project?
Nov 27, 2017
- Timeline for LUAD, UCEC and KIRC projects: given here
Oct 30, 2017: tentative agenda
- Discuss CPTAC-wide use of FireCloud: how to allot funding, make billing projects, add users etc
- Recall supervisor modein FISSFC:
- Update on DSDE collaboration:
- Show recent CGA/DSDE collaboration proposal
- FISS backbone of Jupyter notebooks in FireCloud
- Code generator progress:
- standalone tool
- works on GTEX
- Swagger2 / FireCloud proof of concept has been done
- Full Swagger2 support is next
- Discussion for Wed 11/1 AWG telecon:
- Thoughts omitted from F2F talk, for time constraint: slides 21-39
- Chet: recent CPTAC2 workspaces ... where to go next?
July 2017: FYI on proteomics deliverables from FireCloud CGA team
- FireCloud data workspaces
- one (possibly two - see below) for each of the three CPTAC AWGs (breast, ovarian, colon)
- The workspaces will contain, at a minimum, the end results (protein level quantification) produced by each AWG.
- We may also include the raw MS files, and/or the standardized mzML files. But all of the pipelines used for analyzing these files rely on windows-based software, and so cannot be run on FireCloud.
- We will include the TCGA genomic, clinical and biospecimen data as well - this will help researchers who want to conduct correlative analyses. It will mean, however, that we'll want to create both open and controlled access versions of these workspaces, as the BAMs and VCF files are controlled access.
- We may also include the outputs of the CDAP pipeline, which are published on the CPTAC data portal.
- We will aim to get these workspace in place by the end of August
- Workflows
- Since all of the workflows that run on either the raw MS files or the mzML files (CDAP) include windows-based tasks, they cannot be run on firecloud.
- Mani and Mike's teams are developing workflows for correlative analysis; we agreed to touch base with them at the end of August to see how far along any of these pipelines are and whether they could be included in our deliverables. If not, so be it....I'm hoping that NCI will see the value in the data workspaces for the future development of workflows.
May 31, 2017 On-Site (Broad Institute, Cambridge MA)
- Mike's slides: here
April 4, 2017 Face-to-Face (Bethesda, MD)
- Mike's slides: here